Hi Sander,
Clear about the fact that a new variable is created using derive to report the datetime stop from start and length, but why would you need such variable? For which purpose is this required or in which context?
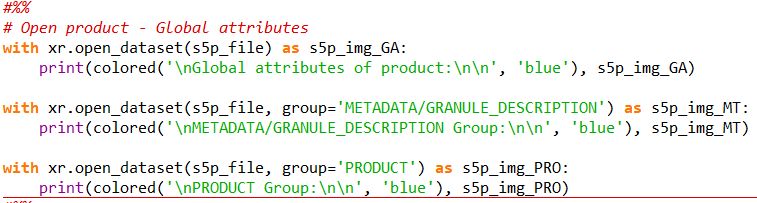
So far, I am testing my methodology in Python with one product only with:
test = harp.import_product(data,operations= “tropospheric_NO2_column_number_density_validity>50;
derive(tropospheric_NO2_column_number_density [Pmolec/cm2]);
derive(datetime_stop {time});
latitude > 42.2 [degree_north] ; latitude < 52.2 [degree_north] ; longitude > -12.2 [degree_east] ; longitude < 22.2 [degree_east];
bin_spatial(1000 ,42.2 ,0.01 ,1000 ,12.2 ,0.01);
derive(latitude {latitude});derive(longitude {longitude});\
keep(NO2_column_number_density,tropospheric_NO2_column_number_density,stratospheric_NO2_column_number_density,NO2_slant_column_number_density,tropopause_pressure,absorbing_aerosol_index,cloud_fraction,sensor_altitude,sensor_azimuth_angle,sensor_zenith_angle,solar_azimuth_angle,solar_zenith_angle)”)
At this point I have my product filtered, converted to molec/cm2, subseted and regridded to my AOI and with new variables (time_stop and the center cordinates of the grid). The output being:
source product = 'S5P_OFFL_L2__NO2____20200105T094546_20200105T112716_11549_01_010302_20200107T025641.nc'
history = "2020-04-01T08:24:43Z [harp-1.9.2] harp.import_product('/shared/Training/ATMO02_MonitoringPollution_Italy/Original4/S5P_OFFL_L2__NO2____20200105T094546_20200105T112716_11549_01_010302_20200107T025641.nc',operations='tropospheric_NO2_column_number_density_validity>50; derive(tropospheric_NO2_column_number_density [Pmolec/cm2]); derive(datetime_stop {time}); latitude > 42.2 [degree_north] ; latitude < 52.2 [degree_north] ; longitude > -12.2 [degree_east] ; longitude < 22.2 [degree_east]; bin_spatial(1000 ,42.2 ,0.01 ,1000 ,12.2 ,0.01); derive(latitude {latitude});derive(longitude {longitude})')"
double datetime_start {time=1} [seconds since 2010-01-01]
float datetime_length [s]
int orbit_index
double sensor_altitude {time=1, latitude=999, longitude=999} [m]
double solar_zenith_angle {time=1, latitude=999, longitude=999} [degree]
double solar_azimuth_angle {time=1, latitude=999, longitude=999} [degree]
double sensor_zenith_angle {time=1, latitude=999, longitude=999} [degree]
double sensor_azimuth_angle {time=1, latitude=999, longitude=999} [degree]
double pressure_bounds {time=1, latitude=999, longitude=999, vertical=34, 2} [Pa]
double tropospheric_NO2_column_number_density {time=1, latitude=999, longitude=999} [Pmolec/cm2]
double tropospheric_NO2_column_number_density_amf {time=1, latitude=999, longitude=999} []
double NO2_column_number_density {time=1, latitude=999, longitude=999} [mol/m^2]
double NO2_column_number_density_amf {time=1, latitude=999, longitude=999} []
double stratospheric_NO2_column_number_density {time=1, latitude=999, longitude=999} [mol/m^2]
double stratospheric_NO2_column_number_density_amf {time=1, latitude=999, longitude=999} []
double NO2_slant_column_number_density {time=1, latitude=999, longitude=999} [mol/m^2]
double cloud_fraction {time=1, latitude=999, longitude=999} []
double absorbing_aerosol_index {time=1, latitude=999, longitude=999} []
double cloud_albedo {time=1, latitude=999, longitude=999} []
double cloud_pressure {time=1, latitude=999, longitude=999} [Pa]
double surface_albedo {time=1, latitude=999, longitude=999} []
double surface_altitude {time=1, latitude=999, longitude=999} [m]
double surface_pressure {time=1, latitude=999, longitude=999} [Pa]
double surface_meridional_wind_velocity {time=1, latitude=999, longitude=999} [m/s]
double surface_zonal_wind_velocity {time=1, latitude=999, longitude=999} [m/s]
double sea_ice_fraction {time=1, latitude=999, longitude=999} []
double tropopause_pressure {time=1, latitude=999, longitude=999} [Pa]
double datetime_stop {time=1} [s since 2000-01-01]
long count {time=1}
float weight {time=1, latitude=999, longitude=999}
float solar_zenith_angle_weight {time=1, latitude=999, longitude=999}
float solar_azimuth_angle_weight {time=1, latitude=999, longitude=999}
float sensor_zenith_angle_weight {time=1, latitude=999, longitude=999}
float sensor_azimuth_angle_weight {time=1, latitude=999, longitude=999}
double latitude_bounds {latitude=999, 2} [degree_north]
double longitude_bounds {longitude=999, 2} [degree_east]
double latitude {latitude=999} [degree_north]
double longitude {longitude=999} [degree_east]
The objecitve is to continue my analysis using xarray so I need to export my harp.Product into an xarray.DataArray. Using:
import xarray as xr
export_path = 'my/path/test6.nc'
harp.export_product(test, export_path,file_format='netcdf')
test_open = xr.open_dataset(export_path)
test_open
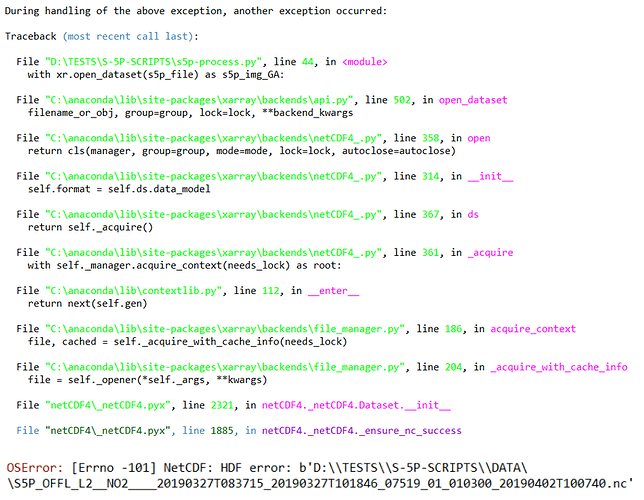
I get the following error:
ValueError: unable to decode time units ‘s since 2000-01-01’ with the default calendar. Try opening your dataset with decode_times=False.
What I am missing on the way I export my product?