Hi,
I am trying to read a Sentinel-5p Level-2 NO2 product using the HARP python interface:
import harp
product = .../S5P_OFFL_L2__NO2____20200401T110350_20200401T124521_12784_01_010302_20200403T040102
test_harp = harp.import_product(product)
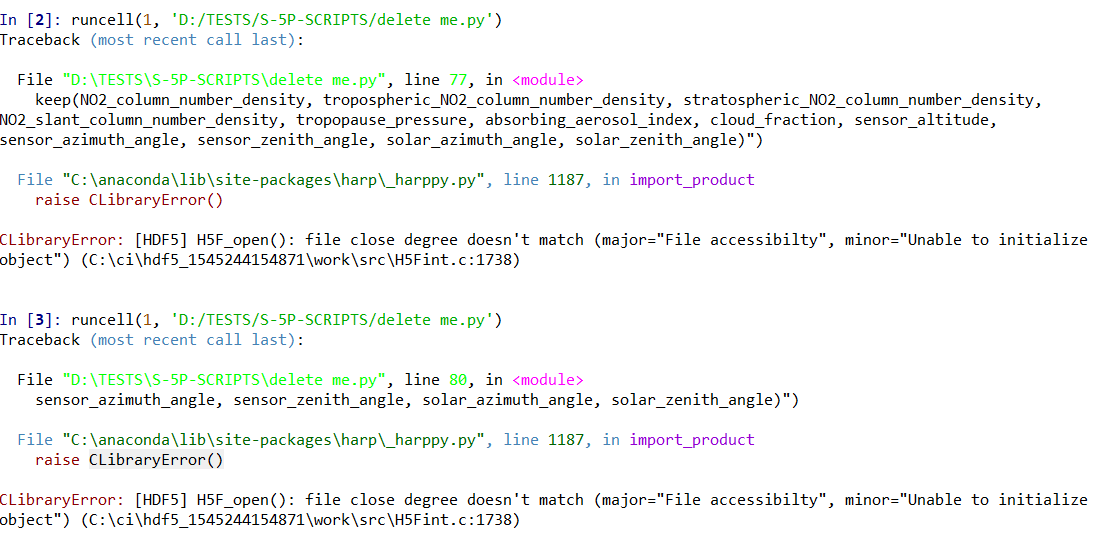
I am getting an error from time to time claiming:
---------------------------------------------------------------------------
CLibraryError Traceback (most recent call last)
~/Training/Sentinel.py in <module>
----> 1 test_harp = harp.import_product(product)
~/anaconda3/envs/rus/lib/python3.7/site-packages/harp/_harppy.py in import_product(filename, operations, options, reduce_operations, post_operations)
1185 if _lib.harp_import(_encode_path(filename), _encode_string(operations), _encode_string(options),
1186 c_product_ptr) != 0:
-> 1187 raise CLibraryError()
1188
1189 try:
CLibraryError: [HDF5] H5F_open(): file close degree doesn't match (major="File accessibilty", minor="Unable to initialize object") (H5Fint.c:1738)
When acessing the file with xarray - xarray.import_dataset(product) any issue in the process happens and I can process my data properly.
Any clue on this matter?