Good evening Sander,
I wanted to ask about the sequence of operations/options in harpmerge, command line edition.
I am using
harpmerge -a ‘SO2_column_number_density>0.8; SO2_column_number_density_validity!& 255;scan_direction_type==0’ -ap ‘bin_spatial(721,-90.0,0.25,1441,-180.,0.25);derive(latitude {latitude}); derive(longitude {longitude});derive(SO2_column_number_density [DU]);keep(latitude, longitude, latitude_bounds,longitude_bounds,SO2_column_number_density,solar_zenith_angle, cloud_fraction,SO2_column_number_density)’ *.HDF5 temp.nc
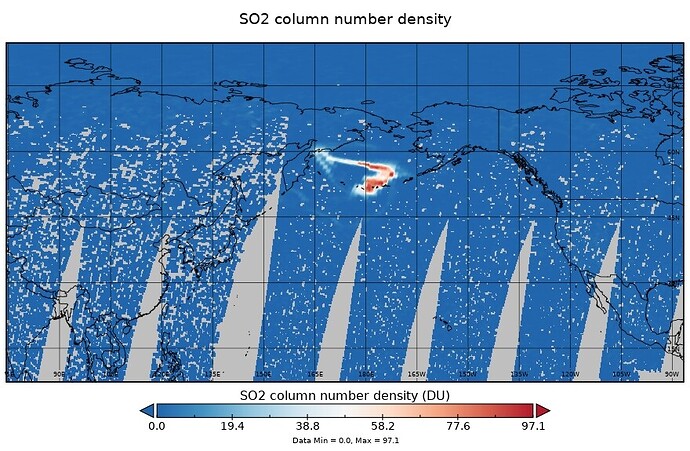
On the Raikoke eruptive day of 23.06.2019, with GOME2C GDP4.9
I want to exclude all pixels with SO2 load of less than 0.8 D.U.
However, I seem to be left with a number of values lower that that. Same if I do not bin_spatial.
There are actual values between 0 and 0.8D.U. in the data, it is not a case of panoply not being able to deal with NaNs or whatever.
What do you think? maybe the sequence of operations has to change?
Furthermore, I just realise that the derive(SO2_column_number_density [DU]) is not needed since this is done automatically for GOME2.
Many thanks
MariLiza