I want to create a harp compliant L2 file (one that is not in the list of ingestions definitions), and would like to ask for some advice. This is a rather general question, but my plan is to do:
1)a mapping of the variable names where possible, according to Variable names — HARP 1.14 documentation.
2)Then adding the needed global and variable attributes (Variable attributes — HARP 1.14 documentation)
3)Then changing the fillvalues to Nan or capture them by valid_min, valid_max, since harp doesn’t recognize FillValue attribute.
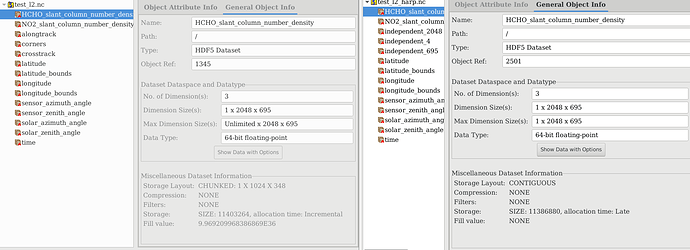
4)Then I dont know how to deal with the dimensions. In the picture below, I have on the left test_l2.nc, which is the L2 that should be (almost) harp compliant and on the right test_l2_harp.nc, which is the output after the command:
harpmerge -f hdf5 test_l2.nc test_l2_harp.nc
Except for the time dimension all dimension in test_l2.nc gets converted to ‘independent_***’ dimensions, which can (by definition) not be flattened.
So, my question here is actually how we can create dimensions that are not converted to independent dimensions by Harp?
The short answer is that you have to squash those dimensions.
You can also see this if you look at how the S5P products are supported by HARP. What is a 3D [time x scanline x ground_pixel] variable in the original product is a single [time] variable in HARP.
So in your case, you should create 1D arrays of length 1423360 (being the time dimension).
The reason why we don’t support arbitrary independent dimensions is because this doesn’t work together with the derivations that we support in HARP (which depend on variables having specific dimensions).
thanks a lot for this quick reply. So, I understand that in order to create a harp compliant L2 file I have to squash variable dimensions (like the number of scanline and ground_pixels) beforehand into the time dimension. Since the command
harpmerge -f hdf5 test_l2.nc test_l2_harp.nc
will only create independent dimensions that cannot be squashed anymore with HARP.
This also implies then, that the command
harpmerge -f hdf5 S5P_OFFL_L2__HCHO___20211001T011729_20211001T025859_20553_02_020201_20211002T165738.nc s5p_test.nc
does squashes scanline x ground_pixels into the time dimension because of the ingestions definitions? So, that Harp recognizes the filename and see it as an s5p product, and that it has to use the appropriate ingestions definition or specific algorithms for s5p files?
I just wonder why there is an independent_4 dimension in the s5p_test.nc (where s5p_test.nc is the output from the above command). This independent_4 dimension appears in the latitude_bounds and longitude_bounds variables (time x independent_4), so how does the derivations in HARP are dealing with this independent dimension?
Exactly. The mapping is also fully described in the last table in the ingestion definitions for these products.
These are intentional independent dimensions. HARP knows how to deal with those for corner coordinates. This is all linked to the variable conversions that HARP can perform with the derive operation. All the supported dimension combinations are described in the Algorithms part of the HARP document, such as, for instance, for latitude. Wherever you see a fixed number (such as ‘2’) or N for a dimension, this is where HARP will expect an ‘independent dimension’.
I wonder when Harp uses an ingestion definition, I thought harp recognized the filename, so for example when the filename starts with ‘QA4ECV_L2_HCHO’ harp uses QA4ECV_L2_HCHO — HARP 1.14 documentation .
However after doing some tests, I could conclude that harp only looks if the id main attribute starts with QA4ECV_L2_HCHO ? Is this the case for all ingestion definitions? Can I also conclude that when the id main attribute does not start with a prefix that is mentioned in Ingestion definitions — HARP 1.14 documentation it will then skip trying to use an ingestion definition?
The logic is that HARP first tries to see if the product is a file matching HARP conventions (based on the Conventions global attribute value. This works for HDF5(/netCDF4), netCDF3, and HDF4 products.
If that doesn’t work, HARP tries an import from a supported external format. It does this based on the coda detection rules (you can see the product class+type of a product with codacheck -V <product>). The combination of product class/type is also included in the tables in the harp ingestion documentation for each supported type of product.
If that also doesn’t work, you will get an error.
For the QA4ECV products specifically you can find the coda detection rules in the associated index.xml file of the QA4ECV codadef.