I am trying now to regridd SO2-COBRA Data with a QA_flag>=0.5. But I don’t know what HARP does with the other values where QA<0.5 and Nan values.
I want to compare model with COBRA SO2. So this is my script:
##### COBRA Daten
import harp
import numpy as np
import matplotlib.pyplot as plt
import sys
from netCDF4 import Dataset
import pandas as pd
import cartopy
import cartopy.crs as ccrs
import xarray
from glob import glob
import matplotlib.cm as cm
import matplotlib as mpl
from scipy.interpolate import griddata as gd
from metpy.interpolate import interpolate_1d
from datetime import datetime as dt
import gc
import datetime
import time
from xarray import DataArray
import dask.array as da
#import xesmf as xe
import h5py
import netCDF4
import harp
sat_data=[]
path_model=glob(‘/work/bd0617/b309194/TROPOMI/final_result/07/final_result20190710_orbit_no_09010.nc’)
n=len(path_model)
print(n)
for i in range (0, 1):
print(i) ***
orbit=path_model[i]***
*** orbit=orbit[75:80]***
*** print(orbit)***
*** time= path_model[i][57:65]***
*** print(time)***
*** try:***
*** path_sat = glob (‘/work/bd0617/b309194/TROPOMI/COBRA_TROPOMI/2019/TROPOMISO2_COBRA_v11_iter4__’+orbit+‘.nc’)***
*** xr_sat = xarray.open_mfdataset (path_sat[0])***
*** f= netCDF4.Dataset(glob(‘/work/bd0617/b309194/TROPOMI/COBRA_TROPOMI/2019/TROPOMISO2_COBRA_v11_iter4__’+orbit+‘.nc’)[0],‘r’)***
*** if ‘QA flag’ in xr_sat:***
*** lon=f[‘longitude_bounds’][:] ***
*** lat=f[‘latitude_bounds’][:] ***
*** so2=f[‘SO2 VCD cobra’][:] ***
*** qa=f[‘QA flag’][:] ***
*** sel=np.where(qa<=1.0) ***
*** f.close()***
*** print(‘Satellite File without group!’)***
*** else:***
*** lon=f[‘Data Fields/longitude_bounds’][:] ***
*** lat=f[‘Data Fields/latitude_bounds’][:] ***
*** so2=f[‘Data Fields/SO2 VCD cobra’][:] ***
*** qa=f[‘Data Fields/QA flag’][:] ***
*** sel=np.where(qa<=1.0) ***
*** f.close()***
*** print(‘Satellite file with group’)***
*** e=netCDF4.Dataset(path_model[i],‘r’)***
*** so2_emac=e.variables[‘vert_col_SO2’][:] ***
*** e.close()***
*** ncfile = Dataset(“/work/bd0617/b309194/HARP_Inputs/07s_days/COBRA_test7_”+str(time)+“_orbit” +str(orbit) +“.nc”, “w”, format=“NETCDF4”)***
*** nc_x = ncfile.createDimension(‘time’, len(sel[0]))***
*** nc_c = ncfile.createDimension(‘corner’, 4) ***
*** nc_lat = ncfile.createVariable(“latitude_bounds”,“f4”,(“time”,“corner”,),zlib=True)#,fill_value=9.96921E36)***
*** nc_lon = ncfile.createVariable(“longitude_bounds”,“f4”,(“time”,“corner”,),zlib=True)***
*** nc_lat[:] = lat[sel]***
*** nc_lon[:] = lon[sel] ***
*** nc_lat.units = ‘degree_north’***
*** nc_lon.units = ‘degree_east’***
*** nc_lat.long_name = ‘latitude of pixel center’***
*** nc_lon.long_name = ‘longitude of pixel center’ ***
*** nc_lon.valid_min = -180. ***
*** nc_lon.valid_max = 180. ***
*** nc_lat.valid_min = -90. ***
*** nc_lat.valid_max = 90.***
*** nc_lon.description =‘longitudes of the ground pixel corners (WGS84)’ ***
*** nc_lat.description = ‘latitudes of the ground pixel corners (WGS84)’ ***
*** nc_so2 = ncfile.createVariable(“SO2”,“f4”,(“time”,),zlib=True) ***
*** nc_so2[:] = (so2[sel]10000) / 2.69e20**
*** nc_so2.units= “DU”***
*** nc_so2.long_name = “SO2 vertical column density” ***
*** nc_so2emac = ncfile.createVariable(“SO2_Emac”,“f4”,(“time”,),zlib=True) ***
*** nc_so2emac[:] = so2_emac[sel] ***
*** nc_so2emac.units= “DU”***
*** nc_so2emac.long_name = “SO2 vertical column density”***
*** nc_qa = ncfile.createVariable(“QA”,“f4”,(“time”,),zlib=True) ***
*** nc_qa[:] = qa[sel]***
*** nc_qa.units= “”***
*** nc_qa.long_name = “Quality Flag” ***
*** ncfile.setncatts({ “Conventions”:“HARP-1.0”}) ***
*** ncfile.close() ***
*** filelist=glob(“/work/bd0617/b309194/HARP_Inputs/07s_days/COBRA_test7_”+str(time)+“orbit" +str(orbit) +“.nc”)***
*** filelistnew=[harp.import_product(ifile,post_operations=‘QA>=0.5’) for ifile in filelist]***
*** post_operations=‘bin_spatial(91,-90.0,2,181,-180.0,2);derive(latitude {latitude});derive(longitude {longitude})’ ***
*** binned=harp.execute_operations(filelistnew,post_operations=post_operations)***
*** harp.export_product(binned, "/work/bd0617/b309194/HARP_Inputs/07s_days/Output_COBRA_test7”+str(time)+“_orbit”+str(orbit)+“.nc”) ***
*** except:***
*** print(‘Fehler’)***
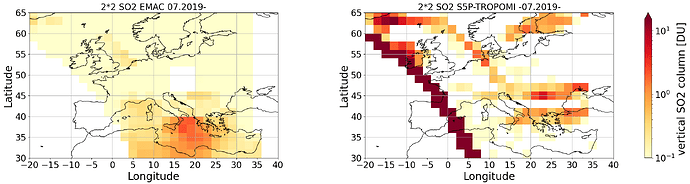
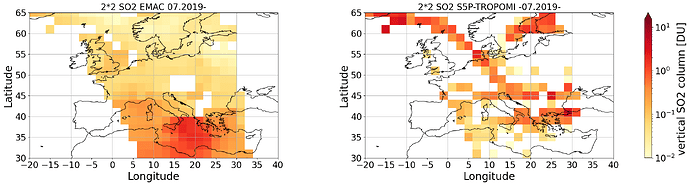
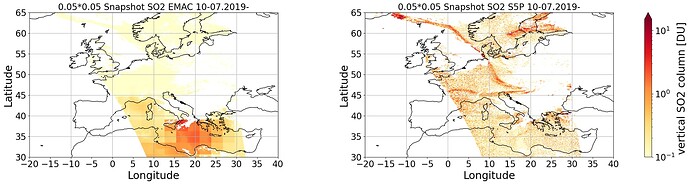
And this is the plot what I got after regridding to a 2*2 Grid:
Thank you in advance for your help.
Ismail Makroum