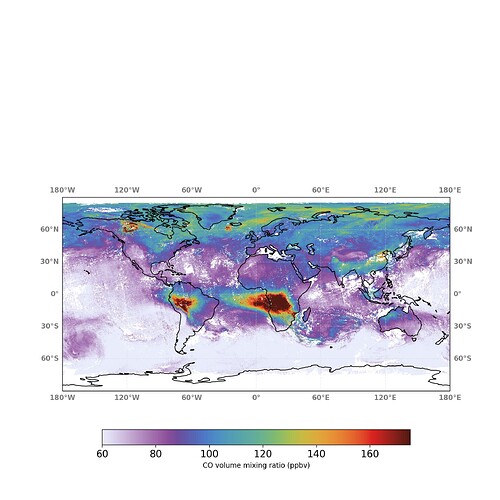
Hello,
I was trying to reproduce a L2__CO map like the one at the website (https://maps.s5p-pal.com/co/).
I first tried averaging data that I downloaded by applying:
basic_operations = ";".join([
"CO_column_number_density_validity>50",
"keep(latitude_bounds,longitude_bounds,CO_column_number_density )",
"bin_spatial(801,-40,0.1,1600,-100,0.1)",
"derive(latitude {latitude})",
"derive(longitude {longitude})",
])
reduce_operations=";".join([
"squash(time, (latitude, longitude, latitude_bounds, longitude_bounds))",
"bin()"
])
s5p_co_merged = harp.import_product(files, basic_operations, reduce_operations=reduce_operations)
but I obtained quite a coarse image. So I have plotted the data provided at the website for those specific days but the image looked still quite different than the website:
I was thinking if the difference could be the use of a 3-days moving average, while I am using the usual time average implemented by Harp. But even in this case, I don’t understand why the data provided by the website look still different when I plot them unless the moving average is applied afterwards.
Also, how is the 3-days moving average applied? Is it considering one day before and one ahead the selected day? And can HARP also perform moving average?
Thank you in advance for the help!
Best,
Serena