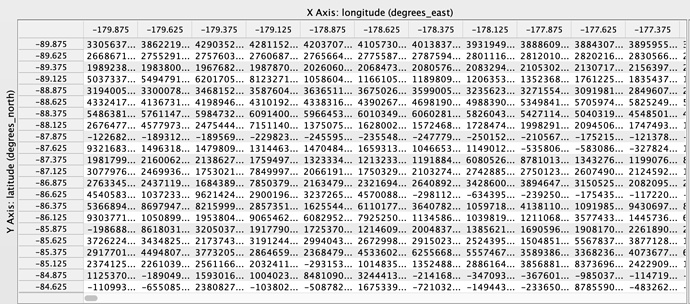
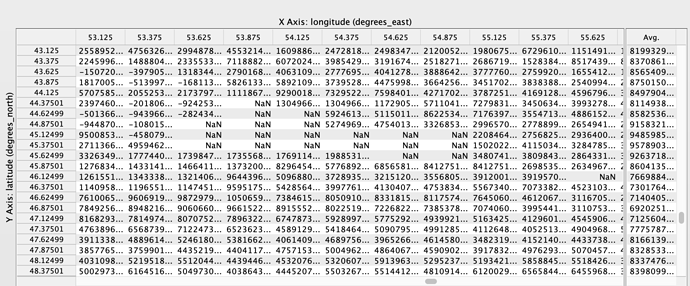
Hi Sander, I’m able to merge the OMI SO2 Data product easily in Python using the following code:
import coda
import harp
import numpy
from os import listdir
#Local variables
productlist = []
Folder = r'/Users/varsharao/Downloads/OMI_HCHO'
OutFolder = r'/Users/varsharao/Downloads/'
#Function for listing files
def list_files1(directory, extension):
return (f for f in listdir(directory) if f.endswith('.' + extension))
#Adding files to a list
FileList = list_files1(Folder, "he5")
for File in FileList:
InFile = Folder + File
pf = coda.open(InFile)
try:
latitude = coda.fetch(pf, '/HDFEOS/GRIDS/OMI_Total_Column_Amount_SO2/Data_Fields/Latitude')
latitude = latitude[:,0]
longitude = coda.fetch(pf, '/HDFEOS/GRIDS/OMI_Total_Column_Amount_SO2/Data_Fields/Longitude')
longitude = longitude[0,:]
so2 = coda.fetch(pf, '/HDFEOS/GRIDS/OMI_Total_Column_Amount_SO2/Data_Fields/ColumnAmountSO2_PBL')
finally:
coda.close(pf)
product = harp.Product()
product.latitude = harp.Variable(latitude, ["latitude"])
product.latitude.unit = "degree_north"
product.longitude = harp.Variable(longitude, ["longitude"])
product.longitude.unit = "degree_east"
product.so2_column_number_density = harp.Variable(so2, ["latitude", "longitude"])
product.so2_column_number_density.unit = "DU"
productlist.append(product)
average = harp.execute_operations(productlist, post_operations="bin();squash(time, (latitude,longitude))")
OutFile = OutFolder + File[19:23] + File [24:26] + ".nc"
harp.export_product(average, OutFile)
However, when I use similar code for the OMNO2d_003 product (NO2 L3) I have to remove lat/long because this isn’t included in the product for some reason and therefore I get the error:
runfile(’/Users/varsha.rao/Dropbox/untitled3.py’, wdir=’/Users/varsha.rao/Dropbox’)
Traceback (most recent call last):
File “/Users/varsha.rao/opt/anaconda3/envs/visan/lib/python3.7/site-packages/harp/_harppy.py”, line 997, in _export_product
_export_variable(name, product[name], c_product)
File “/Users/varsha.rao/opt/anaconda3/envs/visan/lib/python3.7/site-packages/harp/_harppy.py”, line 862, in _export_variable
raise Error(“dimensions missing or incomplete”)
Error: dimensions missing or incomplete
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File “/Users/varsha.rao/Dropbox/untitled3.py”, line 30, in
average = harp.execute_operations(productlist, post_operations=“bin();squash(time)”)
File “/Users/varsha.rao/opt/anaconda3/envs/visan/lib/python3.7/site-packages/harp/_harppy.py”, line 1373, in execute_operations
_export_product(product, c_product_ptr[0])
File “/Users/varsha.rao/opt/anaconda3/envs/visan/lib/python3.7/site-packages/harp/_harppy.py”, line 999, in _export_product
raise Error(“variable ‘%r’ could not be exported (%s)” % (name, str(_error)))
Error: variable ‘‘no2_column_number_density’’ could not be exported (dimensions missing or incomplete)
This is the code I’m running for the NO2 average:
import coda
import harp
import numpy
from os import listdir
#Local variables
productlist = []
Folder = r'/Users/varsha.rao/Downloads/2017_PeakLockdown/'
OutFolder = r'/Users/varsha.rao/Downloads/'
#Function for listing files
def list_files1(directory, extension):
return (f for f in listdir(directory) if f.endswith('.' + extension))
#Adding files to a list
FileList = list_files1(Folder, "he5")
for File in FileList:
InFile = Folder + File
pf = coda.open(InFile)
try:
no2 = coda.fetch(pf, '/HDFEOS/GRIDS/ColumnAmountNO2/Data_Fields/ColumnAmountNO2TropCloudScreened')
finally:
coda.close(pf)
product = harp.Product()
product.no2_column_number_density = harp.Variable(no2)
product.no2_column_number_density.unit = "molec/cm2"
productlist.append(product)
average = harp.execute_operations(productlist, post_operations="bin();squash(time)")
OutFile = OutFolder + File[19:23] + File [24:26] + ".nc"
harp.export_product(average, OutFile)