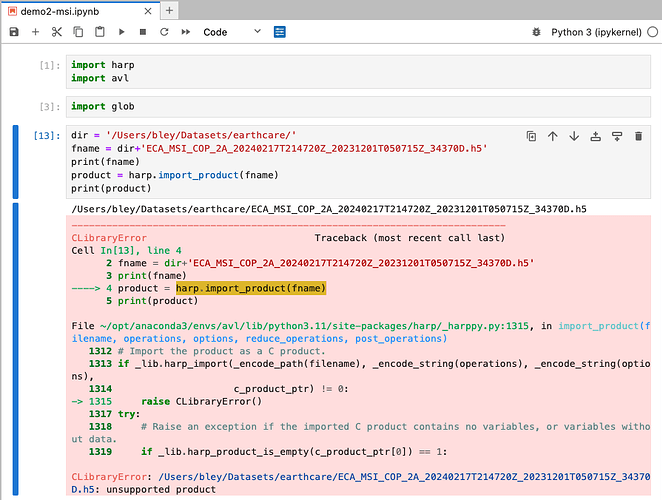
I am facing an issue reading EarthCARE test data downloaded from ESA EarthCARE Online Dissemination from the rehearsal campaign. I use the demo2-msi via Jupyter lab and get the message: unsupported product - see screenshot below. However, the Conda installation of avl worked fine and I can import harp and avl…
This error means that the underlying CODA library that is used for reading couldn’t recognize the product, which is most likely due to the EARTHCARE codadef file not being found. Did you perhaps have a CODA_DEFINITION environment variable set?
This environment variable should either be absent or it should point to the directory where the EARTHCARE .codadef file is located (usually under share/coda/definition in your conda environment).
Thanks for your quick reply.
I have not set a coda def environment variable. I just did this for reading Aeolus, when I physically used coda for reading through python.
Now, I am using the avl conda environment set up as suggested and the EarthCARE coda def file is in the directory as specified by you: /share/coda/definitions/EARTHCARE-20211122.codadef
But I still get the same error, no matter if I try via python, Ipython, or jupyter-lab.
You should not use /share/coda/definitions/EARTHCARE-20211122.codadef. That path doesn’t exist.
You should set the CODA_DEFINITION variable to a path that is relative to your conda environment. e.g. /Users/bley/opt/anaconda3/envs/avl/share/coda/definitions
I now see what is going on. The name of your product is incorrect.
It should follow the naming conventions as defined in ‘EarthCARE Products Definitions,
Volume 1 - Common Products Definitions’:
<Mission_Id>_<File_Class>_<File_Type>_<File_Instance_Id>
For example:
ECA_EXAA_BBR_NOM_1B_20171026T143255Z_20171026T210218Z_10398B
Your product is missing the file class part (EXAA).
Indeed, that was the issue. I must have accidentally changed the file name. Now it works alright. Thanks!