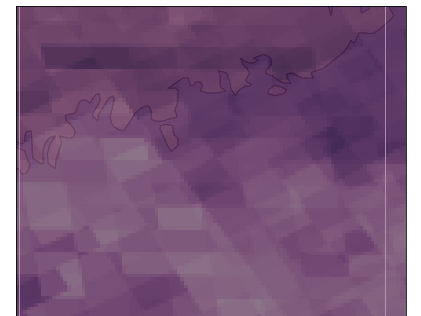
Hello,
I’m trying to plot the time-averaged total column amounts of NO2 over the Eastern Seaboard and I keep encountering the following issue when I plot the data:
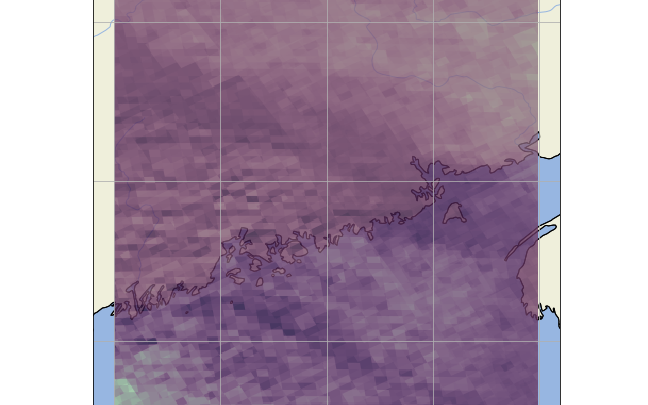
It may be difficult to see, but it appears as though there is seemingly multiple gridboxes overlapping each other, and are not actually merged/averaged into a single gridbox.
Here is my harp import code, there should be only three TROPOMI NO2 files being imported in this example, which will only cover the dates 2018-04-30 to 2018-05-01 (2 days).
I’ve also included the “reduce_operations” argument as well, choosing to merge the files over the time variable (hopefully taking the temporal average).
#### TEST ####
operations = ";".join([
"tropospheric_NO2_column_number_density_validity>0",
"keep(latitude_bounds,longitude_bounds,datetime_start,datetime_length,tropospheric_NO2_column_number_density)",
"derive(datetime_start {time} [days since 2000-01-01])",
"derive(datetime_stop {time}[days since 2000-01-01])",
"exclude(datetime_length)",
"bin_spatial(800, 40, 0.01, 400, -70, 0.01)",
"derive(tropospheric_NO2_column_number_density [Pmolec/cm2])",
"derive(latitude {latitude})",
"derive(longitude {longitude})"
])
reduce_operations = ";".join([
"squash(time, (latitude, longitude, latitude_bounds, longitude_bounds))",
"bin()"
])
harp_L2_L3 = harp.import_product(NO2_files[:3], operations = operations,
reduce_operations = reduce_operations)
export_folder = "{export_path}/{name}".format(export_path=export_path, name=i.split('/')[-1].replace('L2', 'L3'))
harp.export_product(harp_L2_L3, export_folder, file_format='netcdf')
I then plot it using a similar code to that used in “Use case #3”:
NO2_val = harp_L2_L3['tropospheric_NO2_column_number_density'].data
gridlat_NO2 = np.append(harp_L2_L3.latitude_bounds.data[:,0], harp_L2_L3.latitude_bounds.data[-1,1])
gridlon_NO2 = np.append(harp_L2_L3.longitude_bounds.data[:,0], harp_L2_L3.longitude_bounds.data[-1,1])
fig = plt.figure(figsize=(20,20))
ax = plt.axes(projection=ccrs.PlateCarree())
# ax.set_global()
ax.add_feature(cartopy.feature.RIVERS)
ax.add_feature(cartopy.feature.OCEAN)
ax.add_feature(cartopy.feature.LAND, edgecolor='black')
ax.coastlines('10m')
ax.gridlines()
# ax.stock_img()
colortable = cm.tokyo
# # NO2
img = plt.pcolormesh(gridlon_NO2, gridlat_NO2, NO2_val[0],
cmap=colortable, transform=ccrs.PlateCarree(), alpha=0.80)
cbar = fig.colorbar(img, ax=ax,orientation='horizontal', fraction=0.04, pad=0.1)
cbar.ax.tick_params(labelsize=14)
plt.show()
Which gives the aforementioned problem.
So sorry for the interruption, but I would be very grateful for any help or direction!